
Interaction proteomics explores the role of tau in health and disease – Applications of proteomics in neuroscience

Tyler Ford
August 3, 2023
The molecular mechanisms underlying Alzheimer’s disease are poorly understood but are associated with the aggregation of amyloid beta outside neurons and tau inside neurons. How the aggregation of these proteins relates to the underlying mechanisms of the disease is not well understood. Aggregation could be a symptom or a cause. To better understand the roles of tau and beta amyloid in both health and disease, researchers have begun to leverage the incredible power of proteomics.
With specific reference to the tau protein, Tracy, et al recently used interaction proteomics to profile the proteins that wildtype and pathogenic varieties of tau interact with. Their work points to roles for tau in synaptic vesicle trafficking and cellular energetics that may be negatively impacted in Alzheimer’s disease. These findings may enable the development of biomarkers and treatments focused on these pathways.
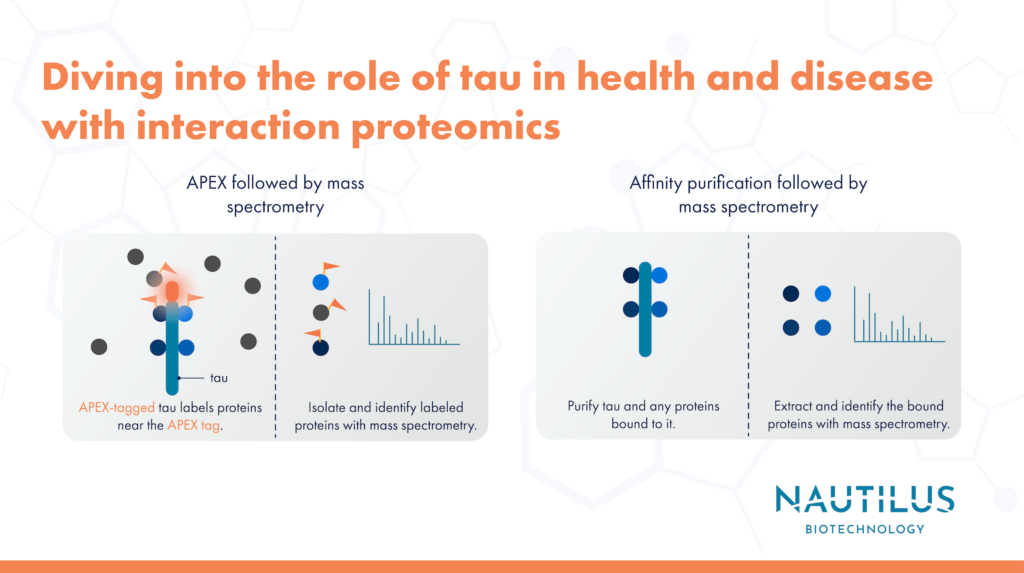
What is interaction proteomics?
With interaction proteomics, researchers label and/or isolate proteins that interact with a protein of interest and profile these interaction partners using techniques like mass spectrometry. Interaction proteomics reveals associations between a protein of interest and other components of a cell’s molecular machinery. Consistent associations between a protein of interest and other proteins with particular functions indicate that the protein of interest is involved in those functions.
Interaction proteomics reveals tau associations with synaptic and mitochondrial proteins
In this work, Tracy, et al differentiated human induced pluripotent stem cells (iPSCs) into neurons and performed two types of interaction proteomics with them:
- APEX labeling followed by mass spectrometry
- Affinity purification followed by mass spectrometry
In the APEX labeling experiments, researchers genetically tagged various forms of tau with the APEX protein in iPSC-derived neurons. Under the proper conditions, APEX will cause biotin molecules to be added to electron-rich amino acids in proteins near the APEX-tagged tau. Using biotin-binding antibodies, researchers purified the biotin-labeled proteins from the cells and used mass spectrometry to identify them. This revealed proteins that were in close enough proximity to APEX-tau to be labeled.
Results from the APEX experiments showed that associations between tau and other proteins change upon neuronal stimulation. For example, the researchers saw increased association with synaptic vesicle proteins upon neuronal stimulation, suggesting that interactions with these proteins may facilitate tau release at synapses and may enable tau to spread between neurons.
These researchers used affinity purification and mass spectrometry to reveal proteins that have longer-lived interactions with tau. Here, they used antibodies to purify tau and any other proteins that were bound to it. Then these binding partners were released and identified via mass spectrometry.
Results from the affinity purification experiments showed that there were differential associations between wild-type tau and mutated versions of tau known to be associated with genetic forms of dementia. Mutant tau proteins interacted less with mitochondrial proteins, and cells producing mutated tau had impaired energetics compared to those producing wild-type tau. In addition, when the researchers looked at the abundance of tau-interacting proteins from mitochondria in people with Alzheimer’s, they discovered that decreases in these interacting proteins were associated with increased disease severity.
These results point to impaired energetics as a possible mechanism of tau-induced neuron dysfunction in Alzheimer’s. Future treatments could be designed to counteract these energetic effects.
Enabling more studies of protein interaction and function with next-generation proteomics
This study demonstrates the incredible power of proteomics to reveal the functions of proteins in both normal biology and disease. While studies like this are currently restricted to a small number of labs where researchers have the expertise to complete them, next-generation proteomics technologies like the Nautilus Proteome Analysis Platform aim to make such work far more accessible. We are designing our platform to enable many more studies like this and provide unforeseen insights into human health that will make it possible to exquisitely target diseases like Alzheimer’s at the molecular level.
Check out our round up of proteomics and neuroscience posts.
Watch this animation to learn how next-generation proteomics technologies can fuel neuroscience.
MORE ARTICLES

