Functional proteomics aims to identify the roles proteins play in carrying out cellular processes at the molecular, mechanistic level. As scientists explore the proteome, the collection of all proteins inside a cell, tissue, or sample, they must figure out not only which proteins are present, but also how those proteins work and interact with other proteins and molecules around them. In essence, functional proteomics helps show scientists exactly what proteins do.
The list of protein functions is long, and still growing. Proteins are responsible for many essential biological functions and processes, from making muscles contract to maintaining cellular structure. They catalyze reactions that would otherwise occur far too slowly, move things around inside cells, and much more. We’re only scratching the surface of the proteome and new proteins and protein functions are continually being discovered!
Functional proteomics is generally conducted using targeted proteomics approaches, or proteomic approaches that set out to study a specific protein or proteins of interest. For example, functional proteomics applications are already showing how the tau protein is involved in Alzheimer’s disease and how specific proteins increase cancer risk. Next-generation proteomics tools like the Nautilus Proteome Analysis Platform are designed to expand these and similar applications even further. Below, we dive deeper into how functional proteomics works and what the future might hold.
Check out the video below to learn how the Nautilus Platform is designed to quantify proteins across the wide dynamic range of the proteome
Examples of functional proteomics
Scientists carry out functional proteomics in a number of ways, but there are two common types of protein analysis associated with the field. These are analyses of protein co-expression and protein-protein interactions.
A protein co-expression analysis identifies proteins that are consistently produced at the same time. If one protein is consistently found alongside another protein with a known function, it is likely that the two proteins are involved in the same process.
Scientists can use traditional proteomics tools like affinity screens and mass spectrometry to conduct protein co-expression analyses. Affinity screens use antibodies and other reagents to bind to specific proteins. Using these reagents, proteomics experiments are often set up so that target binding generates a quantifiable signal that can be used to measure protein expression. Mass spectrometry enables researchers to identify multiple proteins in a sample by breaking them apart and identifying their pieces through their characteristic masses and charges. While a bit more complex than affinity screens, mass spectrometry can also be used to measure protein expression levels. Both of these proteomics techniques can be used to detect proteins that are produced together under various conditions.
A protein-protein interaction analysis examines the ways proteins affect other proteins around them. For example, myosin filaments bind and unbind actin filaments to cause muscles to contract and relax. The binding of one protein to another is a good indication that both proteins are involved in the same function.
Dissecting protein-protein interactions can be a bit more complicated than protein co-expression analyses, but also involves the use of affinity reagents and/or mass spectrometry. In one common proteomics technique, an affinity reagent is used to isolate a target protein from a sample under conditions that do not disrupt the other proteins bound to the target. Researchers can use additional affinity reagents or mass spectrometry to identify the other proteins bound to the target. These protein interaction partners may be involved in the same processes as the target.
These are just a couple of the techniques scientists use to study protein functions, but there are many more. Functional proteomics techniques include fluorescence microscopy, proximity labeling, and some scientists have compared genomes from different organisms to find similar protein-coding sequences and infer protein function. Scientists also use computational methods to predict how an unstudied protein might function based on its structural similarity to other proteins of known function. All that’s to say there is a wide range of methods available to scientists wishing to learn more about protein functions and it’s an exciting time to work in this field!
Listen to the Translating Proteomics podcast for a fascinating discussion of the applications of proteomics
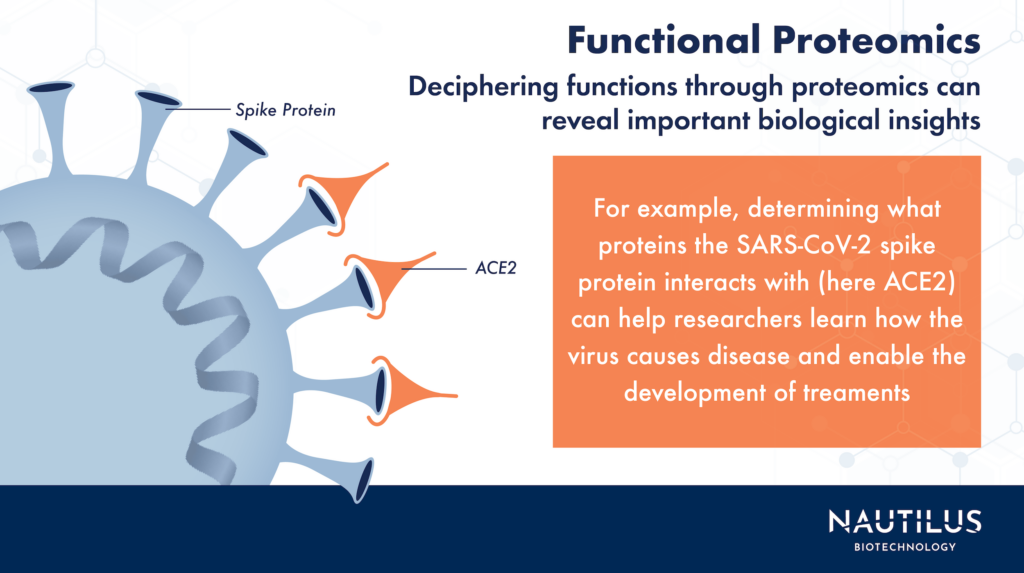
Functional proteomics reveals protein-protein interactions in COVID-19
Functional proteomics is typically used when encountering a new protein, or a protein in a new context. So, it’s ideal for studying an emerging virus with novel proteins, like during the first months of the COVID-19 pandemic. As the virus spread, scientists applied functional proteomics in the race to understand the ways the virus infected cells and replicated. Functional proteomics techniques helped scientists identify viral proteins and their roles in infection.
In a paper from April 2020, published in Nature, researchers used a proteomic analysis based on affinity-purification mass spectrometry to map protein-protein interactions between 26 proteins from the SARS-CoV-2 virus and proteins in the human body. The 26 virus proteins interacted with human proteins with functions like DNA replication, lipid modification, and more. In particular, they found that many of the interacting human proteins were expressed in the lungs, which supported the hypothesis that SARS-CoV-2 predominantly attacks the lungs.
In total, the researchers found 332 protein-protein interactions, 66 of which were druggable with existing compounds. From that list, they found compounds that showed antiviral activity, pointing the way toward potential treatments for COVID-19. Promising compounds included metformin, shown in later trials to help with symptoms of long COVID, as well as Silmitasertib, which helped patients in a small trial recover more quickly. This is just one example of the powerful and actionable findings that can come from fuctional proteomics research.
Next-generation tools for functional proteomics
To understand how proteins interact or are co-expressed in a sample in a comprehensive way, researchers need proteomics technologies that can identify nearly all the proteins in a sample. With next-generation proteomics tools like the Nautilus Proteome Analysis Platform, we expect more than 95% of the proteins of any sample will be accessible to researchers, including both low- and high-abundance proteins. With that level of comprehensive data on the proteome, researchers will hopefully be able to identify more protein interactions, confidently detect more co-expressed proteins, and delineate more protein functions.
MORE ARTICLES


