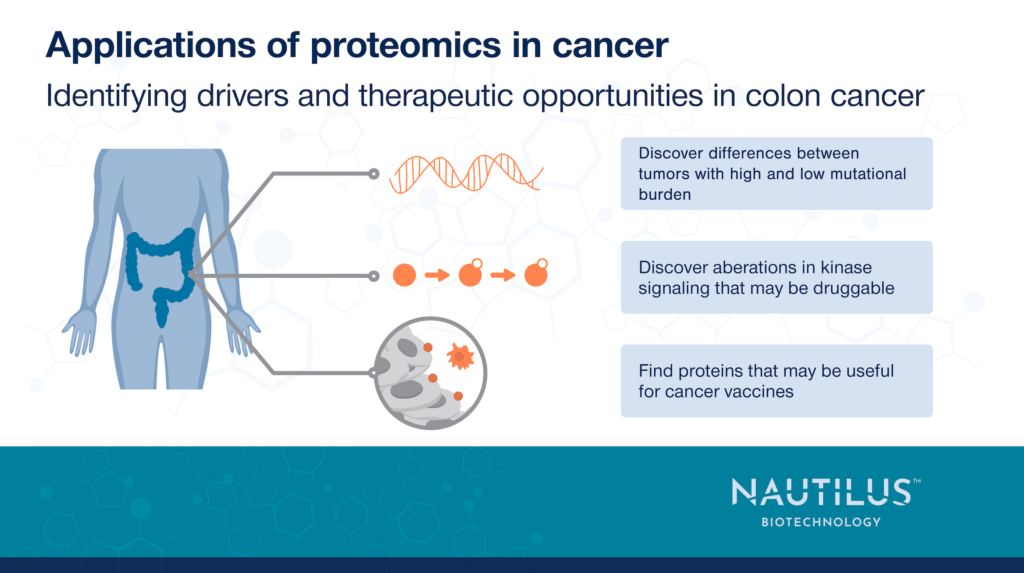
Applications of proteomics in cancer – Identifying drivers and therapeutic opportunities in colon cancer

Tyler Ford
October 24, 2023
Colon cancer is one of the most deadly cancers in the US causing roughly 8.6% of cancer deaths according to the National Cancer Institute. It’s predicted that more than 153,020 people in the US will be diagnosed with this devastating disease in 2023. If caught early, colon cancer can be treated by tumor surgical resection, and new targeted therapies for advanced disease are in development. Nonetheless, given the incredible burden of this disease, it’s clear that we need a better understanding of its molecular underpinnings.
Researchers associated with the Clinical Proteomic Tumor Analysis Consortium have carried out monumental efforts to characterize a variety of tumors using multiomics analyses. Vasaikar et al. 2019 conducted such an analysis on colon cancer tumors in 2019 and compared their genomic, transcriptomic, proteomic, and phosphoproteomic profiles to those of matched adjacent tissues. We cover some of their key findings below and highlight how their work elucidates the biology of various colon cancer subtypes while also identifying points for targeted therapeutic intervention and the development of cancer vaccines. Future work in this area may be accelerated with next-generation proteomics technologies like the NautilusTM Proteome Analysis Platform.
Characterizing colon cancer by genomic mutations and their effects on the transcriptome and proteome
Vasaikar et al. 2019 first divided the tumor samples into two groups – those that were hypermutated and those that were not. The hypermutated group generally contained samples with microsatellite instability – a large number of microsatellite length polymorphisms – and a higher number of mutations overall including single nucleotide variants and indels. In many cases, microsatellite polymorphisms likely resulted from mutations in mismatch repair genes which are known to cause microsatellite instability.
Overall, the hypermutated and non-hypermutated groups had frequent mutations in different sets of genes indicating that they had different etiologies. Investigating the frequently mutated genes provided clues as to the mechanisms underlying tumor growth.
Hypermutated colon tumor samples often had mutations in DNA repair enzymes as well as high frequency mutations in 9 additional genes. These genes included CASP5, which encodes a protein involved in programmed cell death and RNF43, which encodes a protein that regulates cell growth pathways.
Non-hypermutated tumors made up most samples and had a high frequency of mutations in a different set of genes that included:
- APC – a protein that plays a variety of roles in regulating cellular proliferation and is a well-known tumor suppressor.
- TP53 – also a known tumor suppressor.
- SOX9 – SOX9 was particularly interesting because it was both frequently truncated at the gene-level and over-expressed at the protein level. Gene truncation often leads to non-functional proteins, and a high level of truncation in tumor cells would usually indicate that a gene encodes a tumor suppressor. Yet, the high levels of SOX9 protein observed here indicate that the truncated protein acts to promote tumor cell proliferation.
Analysis of copy number alterations across colon cancer genomes identified correlations between genomic, transcriptomic, and proteomic alterations. Genes with correlated changes across all these levels were prioritized as drivers of colon cancer and were found to be associated with particular biological pathways. For instance, 6 of the 90 genes prioritized as potential drivers through this multiomic analysis were involved in endocytosis. This process can impact, among other pathways, growth signaling and metastasis and is a promising therapeutic target.
View our animation to see how next-generation proteomics can fuel cancer research
The role of phosphorylation in colon cancer signaling pathways
Kinase signaling pathways can drive tumor growth and the multiomic work here identified some of the kinase pathways at work in colon cancer. For example, phosphoproteomic analysis revealed that the retinoblastoma (RB) protein, which is normally considered a tumor suppressor, was surprisingly upregulated in both gene copy number and in protein expression. Phosphosite analysis revealed that, in many cases, the up-regulated RB protein was more highly phosphorylated in tumors than normal adjacent tissues, and the researchers identified CDK2 as the likely kinase responsible. Further analysis revealed pathways through which phosphorylated RB could both activate tumor cell proliferation and inhibit apoptosis. This work suggests CDK2 inhibition as a potential means of treating colon cancer.
Beyond RB, Vasaikar et al. 2019 identified many additional proteins and phosphosites that were either upregulated or downregulated in tumor tissues. While more research is necessary to parse out the many ways these proteins may be involved in disease, some of them had up-regulated expression that was relatively restricted to tumors. This is an exciting finding because cancer vaccines, which prime the immune system to recognize cancer cells, require such tumor specific proteins, and researchers may be able to use these proteins in future colon cancer vaccines.
Colon cancer subtypes based on genomics, transcriptomics, and proteomics
Vasaikar et al. 2019 combined genomic, transcriptomic, and proteomic data to delineate three “Unified multiomics subtypes.” These subtypes were broadly associated with:
- Microsatellite instability (MSI) – These tumor samples had microsatellite instability, were associated with a previously identified proteomic colorectal cancer subtype (ProS-B, Zhang et al. 2014), and were associated with a previously identified transcriptomic colorectal cancer subtype (CMS1, Guinney et al. 2015).
- Chromosome instability (CIN) – These tumors had high chromosome instability which could lead to alterations in chromosome number, chromosomal rearrangements, and gene copy number changes. They were also associated with a previously identified proteomic colorectal cancer subtype (ProS-E, Zhang et al. 2014) and a previously identified transcriptomic colorectal cancer subtype (CMS2, Guinney et al. 2015).
- Epithelial to mesenchymal transition – These tumor samples had multiomic signatures indicative of the epithelial to mesenchymal transition, a process whereby tumor cells gain the ability to invade other tissues. They were also associated with a previously identified proteomic colorectal cancer subtype (ProS-C, Zhang et al. 2014) and a previously identified transcriptomic colorectal cancer subtype (CMS4, Guinney et al. 2015).
Importantly, tumors from different subtypes are potentially susceptible to different kinds of treatment. For example, tumors in the MSI subgroup have a high level of immune cell infiltration. This indicates that MSI tumors may be susceptible to treatments that enhance the ability of immune cells to kill tumor cells.
Applying findings from the proteogenomic analysis of colon cancer to future studies with next-generation proteomics
The findings from this combined genomic, transcriptomic, and proteomic analysis are highly actionable. They are likely to spur further research both into the roles of the up and down-regulated pathways in colon cancer progression and the potential therapeutic strategies identified. Importantly, this additional work will require higher sample numbers and throughput to validate biological and therapeutic hypotheses. Accordingly, these efforts will be greatly aided by next-generation proteomics technologies, like the NautilusTM Proteome Analysis Platform, that are designed to be more accessible to more researchers in more labs. We hope researchers can use our platform to advance studies like this one and aid in the fight against devastating diseases like colon cancer.
MORE ARTICLES