
The understudied proteins initiative – A new opportunity for functional proteomics

Tyler Ford
September 19, 2023
It may seem hard to believe, but biologists can have a tendency to study the same tried and true subjects. The phenomenon is easily seen in the study of animals, where attention (and funding) is disproportionately given to species referred to as “charismatic megafauna” — the pandas and panthers of the world — when compared to little-known, less marketable species.
Unfortunately, this same problem plagues protein research. As researchers seek to understand all the proteins found in living cells and their biological functions — the proteome — the most well-known proteins get consistent attention, while countless others have barely been annotated at all. What secrets of the proteome could be revealed if attention was turned to the millions of understudied proteins and proteoforms?
“Each deeper insight into an understudied protein can exponentially increase our understanding of biology,” says Parag Mallick, Nautilus Founder and Chief Scientist. “Emerging technologies make it more tractable than ever to study these important proteins.”
The protein annotation gap
Countless proteins have been discovered, but not yet annotated. We know they exist, but their biological functions have not been revealed. In a recent comment in Nature Methods, an international team proposed the formation of an Understudied Protein Initiative to combat this problem and reduce the “annotation gap.”
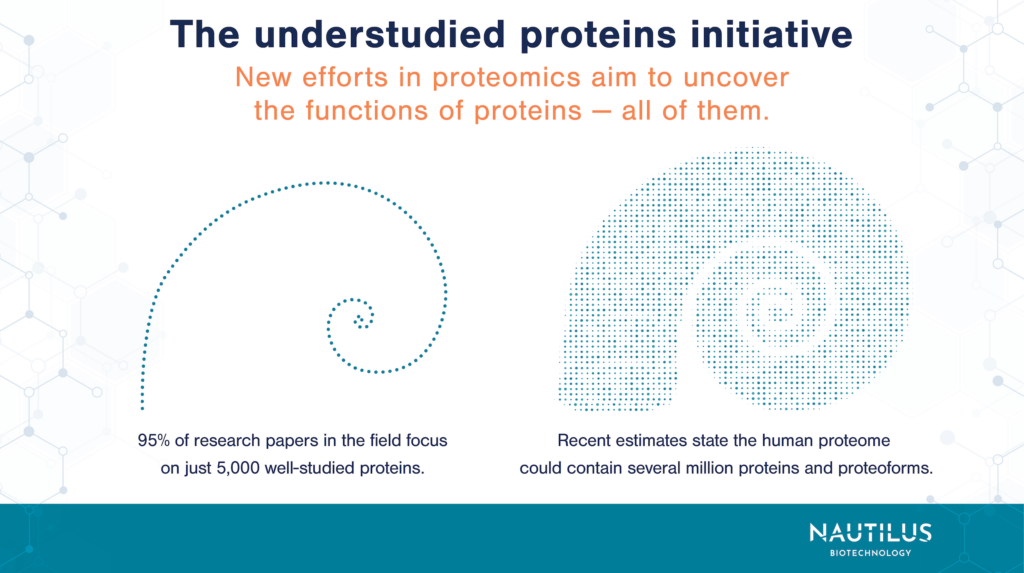
The team points out that the annotation gap is huge: 95% of research papers in the field focus on just 5,000 well-studied proteins. Compare this to recent estimates that the human proteome contains millions of proteoforms.
What is functional proteomics?
The massive gap between the number of proteins and those that have been studied is particularly slowing a field of research called functional proteomics. Functional proteomics studies what roles proteins play inside cells or organisms. Knowing a protein’s function is crucial to understanding its importance for an organism as a whole and can reveal new mechanisms behind diseases and other biological conditions.
Why protein research has stepped further into functional proteomics
There are many reasons why researchers stick to well-known proteins, explain the authors. Some have to do with convenience: It’s easier to study known proteins because the tools and methods to study them, like antibodies or reference data, are readily available. It’s also easier to study proteins that are larger in size, and/or are more abundant in samples. The authors point out that 40% of the least-well-annotated proteins are smaller than 15 kDa.
Some reasons are likely more subconscious. Already-studied proteins might simply be assumed to be more important. There’s also a good deal of inertia that occurs in any research program — it’s hard for researchers to switch gears when everything from lab materials to one’s publication record on a narrow topic are so well-established.
Finally, the entire system in which research occurs is not often set up to encourage bold exploration, even in a cutting-edge field like proteomics. The need for consistent publications and grants makes the risk of exploring the unknown fraught. It can be a challenge to find support for exploratory science like functional proteomics since preference from funders is often given to hypothesis-driven work. Granting agencies prefer clear-cut testing of specific theories over projects asking, “what does this protein do?”
Moving into the understudied proteome
One important application of researching understudied proteins is drug development. Current estimates hold that around 3,000 proteins are druggable, but only 5-10% of these are targeted by pharmaceuticals approved by the FDA. There could be treatments for cancer, autoimmune diseases, and more hidden in that other 90%.
Likewise, there are likely a vast number of biomarkers for disease that have yet to be discovered. Protein biomarker discovery will improve diagnosis and means of monitoring disease progression. For example, brain acid soluble protein 1 (BASP1) was recently identified by quantitative proteomics as a potential biomarker for pancreatic cancer prognosis.
The applications of increasing the number of annotated proteins extend well beyond the biomedical field. Unlocking more of the proteome will lay a foundation for new insights across all of biology. Improving our understanding of cellular processes could enable advances in agriculture, boost our understanding of the natural world, and even inspire new biotechnologies aimed at solving the world’s problems.
New technologies to explore the understudied proteome
Scientists need new technologies to help them explore the vast expanse of the understudied proteome. As Nautilus co-founder and chief scientist Parag Mallick said recently:
“Emerging technologies that make it possible to easily, sensitively and comprehensively measure the proteome will unlock our ability to more effectively understand how every protein contributes to defining biological function and dysfunction, enabling huge acceleration in areas like drug development.”
Indeed, next-generation proteomics platforms may help scientists avoid protein research biases of the past. Technologies like the Nautilus Proteome Analaysis Platform are designed to provide scientists with a broad view of substantively all the proteins in the proteome through discovery proteomics and broadscale protein analysis. The Nautilus Platform is also designed to enable scientists to identify proteoforms through targeted proteoform studies. This will hopefully empower scientists to see much more of the proteome at once and rapidly identify changes in understudied proteins that may be associated with biological functions or disease.
Functional proteomics for the future
The goal of functional proteomics is to understand the functions of proteins — all of them. But with millions of proteins and proteoforms in the human proteome alone, this can seem like an insurmountable task. With the Nautilus Proteome Analysis Platform, we aim to help researchers study the proteome efficiently and effectively. The platform is designed to contain billions of single-molecule protein landing pads that will help scientists understand the full proteome at the resolution of individual proteins. In addition, we’re designing the platform with an integrated and accessible workflow that will enable researchers to achieve biological insights efficiently. With new proteomics tools like ours on the horizon, understudied proteins may soon be a thing of the past.
MORE ARTICLES
