A proteoform is any variant of a genetically encoded protein. Each proteoform is defined by its full set of modifications whether those modifications come from alternative splicing, post-translational modification, or any other source. Thus identifying proteoforms is greatly aided by single-molecule analysis of full-length proteins. Proteoforms may have altered structure, function, interactions, or even solubility, and there are exponentially more potential proteoforms than there are proteins. Below we dive deeper in the mechanisms that give rise to proteoforms, how they can be detected, and how next-generation proteomics platforms with single-molecule analysis capabilities can enable targeted proteoform studies.
Watch this animation to learn how we measure proteoforms on the NautilusTM Proteome Analysis Platform
Processes that give rise to proteoforms
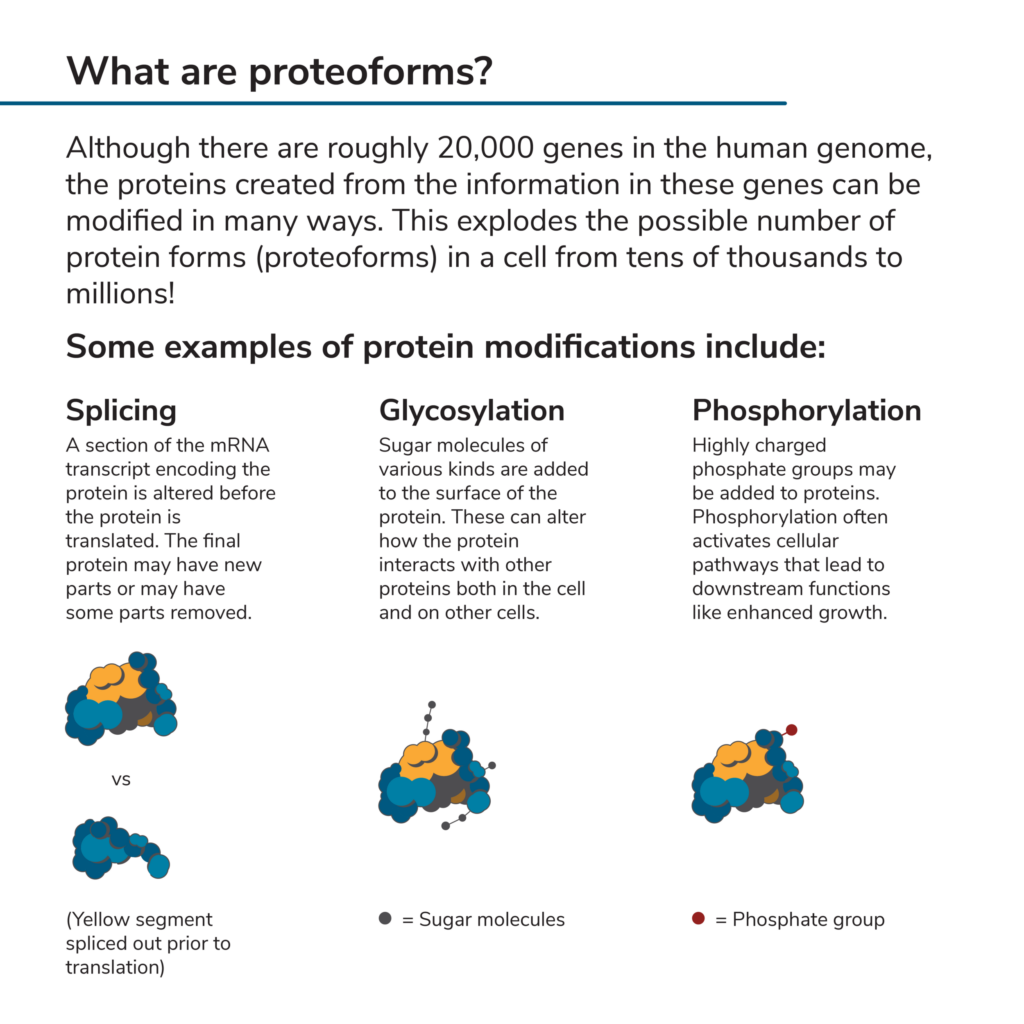
You may have heard that there are roughly 20,000 genes in the human genome. One might assume that, since genes encode proteins, there can be no more than 20,000 different proteins in the human proteome. However, this is far from the case. In the process of transcribing the information in a gene and later translating it, numerous biological pathways can modify a protein. These create diverse versions of the protein known as proteoforms. Some of the ways proteoforms are made include:
- The transcription process may begin at different places in a gene
- Segments of a gene may be edited out
- Transcription may end prematurely resulting in truncation of a protein
- Once this information is used to create a protein, the protein itself can undergo post-translational modification with a variety of attachments such as small molecules, sugars, and even other proteins
All this rearranging and modification explodes the number of possible proteoforms that could theoretically be produced in a human cell from 20,000 to millions (Aebersold et al 2018). Cataloging them all is the goal of the Human Proteoform Project, which has covered nearly 6,000 human proteins and more than 60,000 human proteoforms to date.
Difficulties detecting proteoforms
The modifications that make up proteoforms can drive biologically interesting changes to protein function. Indeed, the precise mix of proteoforms in a cell can have great impacts on cell processes, organ function, and total body systems. Thus, knowing more about the proteoforms present in healthy and diseased cells may give scientists great insights into how cells and tissues operate. Unfortunately, most proteomic analysis technologies used today cannot distinguish proteoforms, can only see a small fraction of them, or do not have the resolution to capture multiple protein modifications and their precise composition within a sample.
- Low sensitivity may limit the proteoforms that can be observed: There are many different ways proteins can be modified to create a wide array of proteoforms. However, for any given cell, it’s possible that only a few proteoforms will be present in high enough abundance to detect them with current methods. There may be low abundance proteoforms that have functional impacts on the cell but they will be hard to see. Tyrosine phosphorylation is a good example. It is known to be extremely important in some proteins but overall levels of tyrosine phosphorylation are very low in most samples.
- Limitations due to protein digestion: On standard proteomic analysis platforms, full length proteins are often digested into small peptides to facilitate analysis. These peptides are identified, and analysis software makes assumptions when piecing together full proteins from the identified peptides. This makes it difficult to map which individual proteins have modifications and instead provides a bulk report of modifications in aggregate. Additionally, not all the peptides that make up a full protein will be observed with most proteomics platforms. Some peptides will be left out of the proteomic analysis and those peptides could have modifications that are missed.
- Bulk measurements: When affinity reagents are used to detect specific types of modification, they are typically used to analyze bulk samples of many proteins at once and individual proteins cannot be resolved. Thus it can be difficult to detect whether individual proteins are modified in multiple ways and to what extent they are modified. For example, it may be difficult to determine if a sample contains three sets of a single protein species modified in three separate ways or one set with three modifications on each protein molecule.
The benefits of detecting proteoforms with single-molecule precision
There are many unknowns when it comes to the world of proteoforms. Scientists have made theoretical predictions about the number of proteoforms that could possibly exist, but it is not at all clear what fraction of these proteoforms are actually made, how they might be distributed across cells, and what functional consequences they have.
Nonetheless, we do know that some protein modifications are highly consequential. For example, the addition of methyl groups to proteins that scaffold DNA can turn off genes that would otherwise suppress cancer development and similar modifications altering gene expression are associated with various cancer outcomes (Nebbioso et al 2018).
With the ability to identify specific proteoforms at the single-molecule level in cells and tissues, scientists can more confidently associate a given proteoform with a given state of health or disease. Such research may lead to better protein biomarkers that more accurately indicate when a person has a particular disease or the identification of specific proteoforms that may make better targets for novel drugs.
Learn more about the importance of understanding proteoforms.
What are proteoform families?
A proteoform family is the complete set of proteoforms derived from a single gene (Shortreed et al., 2016). Thinking about proteoform families helps researchers establish what they should be looking for when searching for proteoforms in cells. That is, thinking in terms of proteoform families can narrow the search parameters used to identify proteoforms experimentally. For instance, when identifying proteoforms from their intact masses and lysine counts as determined by mass spectrometry (see Shortreed et al., 2016), researchers might first identify a single proteoform from a family and then, based on known and possible modifications, identify all the mass signals from other proteoforms that could be derived from that initial proteoform. Rather than identify each proteoform independently, the known and potential relationships between proteoform family members help researchers identify many proteoforms in their data. This may make it easier to experimentally determine differences in proteoform abundance between samples and associate proteoforms with functions.
Enhancing proteoform detection with the Nautilus Proteome Analysis Platform
At Nautilus, we’re developing a proteomic analysis platform that is designed to make it easier to identify proteoforms with increased accuracy and precision.
- The Nautilus Proteome Analysis Platform analyzes single protein molecules in isolation: This gives the platform high sensitivity and thus more potential to detect relatively rare, but possibly important, proteoforms. Single-molecule analysis also makes it possible to determine the extent to which individual proteins are modified and overcomes the problems of bulk measurement
- The Nautilus Proteome Analysis Platform analyzes intact proteins: The platform does not identify proteins from peptide data, so there are no assumptions about how peptides might map to an intact protein. With intact protein analysis, it is theoretically possible to identify modifications across the entire length of a protein and map the precise composition of individual proteoforms within a sample.
By getting a more in-depth view of the proteoforms that exist across samples, we’ll move toward a deeper understanding of how proteoforms impact health and disease. Once scientists observe differences in proteoform abundance across samples, they can investigate whether those differences are functionally significant. The Nautilus Proteome Analysis Platform aims to bring us a long way toward accomplishing these goals.
MORE ARTICLES