When talking about the revolutionary proteomics work we hope to enable with the NautilusTM Proteome Analysis Platform, we think about two general research modalities:
- Broadscale proteomics
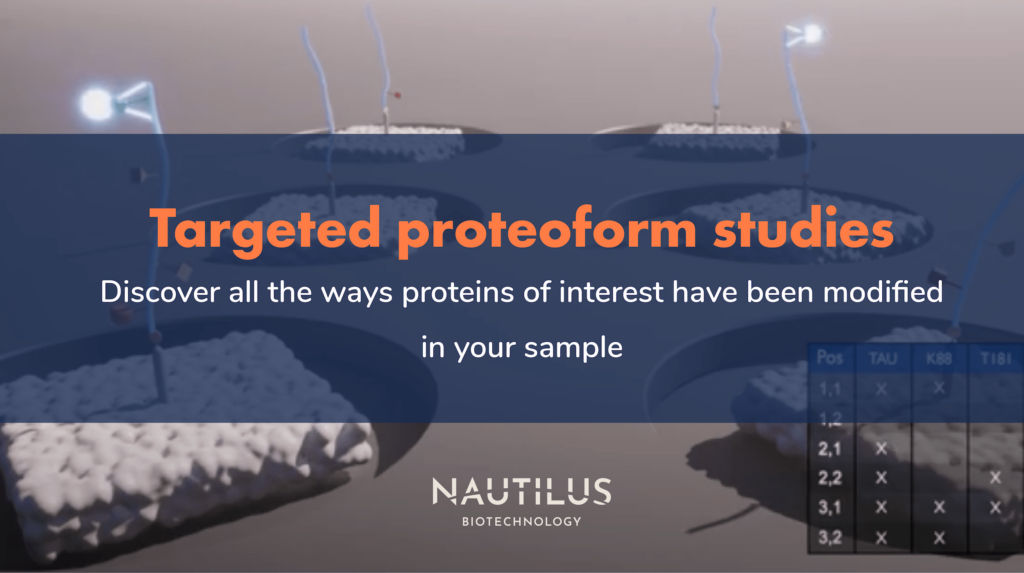
- Targeted proteoform studies
Targeted proteomics generally involves analyzing a subset of specific proteins in the proteome. Efforts like these are most useful when researchers have a protein of interest in mind and want to learn more about how it works inside a cell or organism.
Targeted proteoform studies are a type of targeted proteomics in which researchers zoom in on a particular protein or set of proteins and quantify their many possible variants known as proteoforms. Each proteoform is defined by the full set of modifications it has, whether those modifications come from alternative splicing, post-translational modification, or any other source. Thus identifying proteoforms is greatly facilitated by single-molecule analysis of full-length proteins.
Broadscale proteomics, on the other hand, is an approach that provides researchers with a snapshot of protein identities and abundances across the entire proteome. Both modalities are built upon our single-molecule analysis capabilities and are designed to generate novel and impactful biological insights. Both are necessary to ensure the proteomics revolution bears fruit.
In this blog post, we cover the technical aspects of targeted proteoform studies on the Nautilus Proteome Analysis Platform. We also describe the overarching problems expected to be solved by the platform and discuss a few ways targeted proteoform studies may be applied in future research efforts. In a companion blog post, we cover broadscale proteomics.
Watch this animation for a visual deep dive into proteoform studies on the Nautilus Platform
Targeted proteoform studies on the NautilusTM Proteome Analysis Platform
When performing targeted proteoform studies, researchers generally want to quantify all the proteoforms or a specific subset of proteoforms of their protein of interest. Today, people can look at a single or small handful of protein modifications in bulk with affinity reagents or they can investigate full proteoforms with top down mass spectrometry.
For proteoform analysis on the Nautilus Platform, we use Iterative Mapping. Applying this method to proteoforms, the platform repeatedly interrogates billions of single, intact protein molecules on our massive, nanofabricated protein arrays using off-the-shelf probes designed to bind protein features such as post translational modifications and isoform-specific sequences that combine to define proteoforms. Our machine learning-powered algorithms then use the observed binding patterns to identify each proteoform at the single-molecule level, and identifications are summed to provide proteoform counts.
Targeted proteoform studies are expected to follow a similar workflow to broadscale proteomics on the Nautilus platform. First, a researcher would isolate proteins (or a subset of proteins) from their sample of interest and functionalize the proteins for automated deposition on our 10-billion landing pad arrays. However, instead of flowing multi-affinity probes over the array as in broadscale proteomics, the instrument would flow protein, isoform, and modification-specific probes over the array to identify landing pads containing their proteins of interest and the ways each single protein molecule has been modified – i.e. identify the specific proteoforms found at each landing pad.
Learn more about the importance of understanding proteoforms.
Solving challenges in targeted proteoform studies
One of the major difficulties in targeted proteoform studies on other platforms lies in distinguishing individual proteins modified in multiple ways from multiple proteins modified in separate ways. This is because other proteomics platforms often make bulk measurements or break proteins into peptides and thereby lose data on individual, intact proteins. In contrast, the Nautilus platform is designed to enable researchers to probe single, intact proteins. Thus, researchers can determine the extent of modification to single protein molecules and get an accurate understanding of individual proteoform quantities in a sample.
Applications of targeted proteoform studies
Different proteoforms derived from a single gene can function in very different ways. For example, many cellular signaling pathways are activated by phosphorylation. Abberant activation of such pathways can lead to increased cell growth and give rise to cancer. In another example, tau protein hyperphosphorylation is known to be associated with the progression of Alzheimer’s disease. Looking to more basic biology, in both heath and disease, modifications to histones scaffolding DNA can change how genes are expressed. The collection of proteoforms in a cell can thereby have huge impacts on gene expression and cellular function. Measuring proteoforms at high resolution can thus lead to the identification of novel biomarkers as well as new avenues in drug development.
Indeed, researchers in the Kelleher lab at Northwestern University recently used top-down proteomics (a type of mass spectrometry-based proteomics) to identify proteoforms present in five different tissues. Many of the proteoforms detected appear to relate to the functions of these specific tissues. This kind of work is very challenging now and often requires customized workflows and instrument set-ups but could become far more accessible with new technologies like the Nautilus platform. Similar targeted proteoform studies are expected to lead to a much better understanding of the ways proteins dictate cellular function. This kind of knowledge is essential for the development of novel drugs that target dysfunctional biological pathways specific to certain tissues or organs.
Listen to this episode of our Translating Proteomics podcast for fascinating conversation on the power of proteoforms with Neil Kelleher:
An exciting future for targeted proteoform studies
We hope to make targeted proteoform studies accessible to researchers in many different fields through the Nautilus Proteome Analysis Platform. These studies should complement broadscale proteomics to provide researchers with a clear picture of the many different proteoforms in their biological samples. We expect such understanding to enable scientists to find new biomarkers for disease, develop novel drugs, create a new paradigm in precision medicine, augment biological functions for biotechnological purposes, and find new ways to approach existential crises facing humanity.
Check out some of our other blog posts for more on the fascinating applications of proteomics.
MORE ARTICLES