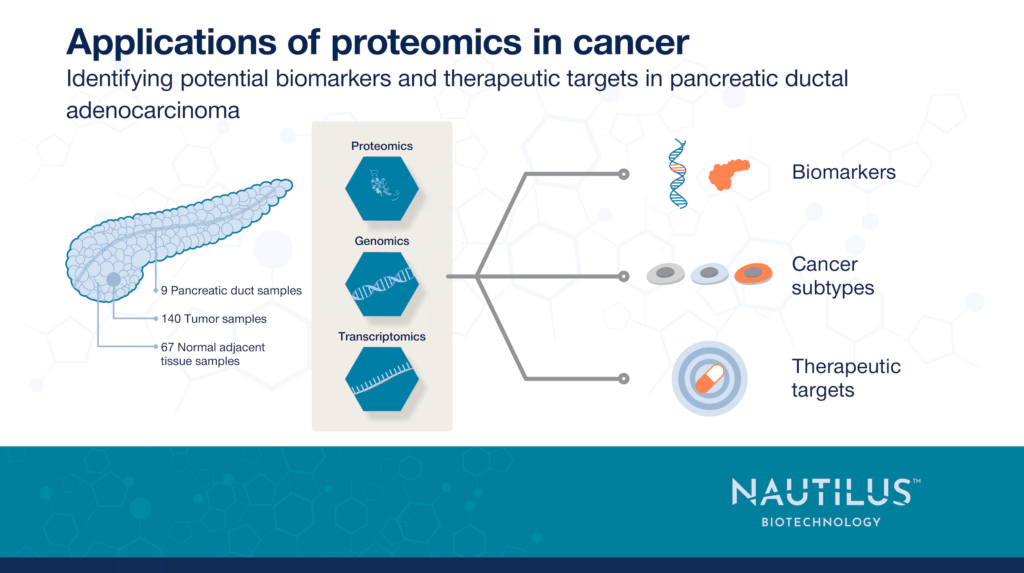
Applications of proteomics in cancer – Identifying potential biomarkers and therapeutic targets in pancreatic ductal adenocarcinoma

Tyler Ford
October 10, 2023
Pancreatic ductal adenocarcinoma (PDAC) is projected to become the second leading cause of cancer deaths by 2030 and is currently the third leading cause of cancer deaths in the US. The disease has a five-year survival rate below 10% and has been difficult to treat because of a lack of early symptoms, reliable screening methods, and early detection tools.
Past genomic studies on PDAC have identified somatic mutations in genes such as KRAS, TP53, CDKN2A, and SMAD4. KRAS activating mutations are the most prevalent genetic alteration in PDAC. However, despite recent successes with the G12C KRAS mutation, KRAS is generally considered difficult to target. Other studies have identified tumor-specific therapeutics that target only a small subset of pancreatic cancers leaving many people without treatment options. While studies that use genomics or transcriptomics can identify signaling pathways involved in PDAC, they alone fall short in fully revealing the connections between genes, transcripts, and proteins.
To overcome these challenges, Cao et al. 2021 examined PDAC using a multiomics approach. They identified potential biomarkers and therapeutic targets for PDAC that may have gone undetected in more limited studies of individual omes. More studies like this one will hopefully become possible if next-generation proteomics technologies are broadly available and accessible. We’re designing the NautilusTM Proteome Analysis Platform to become one such game-changing technology and hope it will drive impactful insights into cancer like those discussed below.
Unraveling the proteogenomic landscape of PDAC
Cao et al. 2021 analyzed pancreatic tumor samples, paired NATs (normal adjacent tissues), and normal pancreatic duct tissues. These samples were of particularly high quality because they were all frozen in liquid nitrogen within, on average, 20 minutes of collection to ensure post-translational modifications could be analyzed accurately. This is incredibly important in a disease like cancer where disruptions to all sorts of signaling pathways involving various proteoforms could result in different prognoses.
This study amassed an immense amount of data including tens of thousands of transcripts, proteins, phosphosites, and glycopeptides. This is a lot of data, but by leveraging information from previously identified mutations (ex: KRAS), the scientists were able to begin homing in on molecular details that only a multiomic approach could capture.
Below, we highlight a few important findings that illustrate the benefits of this multiomic approach.
Linking genomic mutations to functional impact
Genome content doesn’t necessarily correlate with transcript abundance or protein expression, and it doesn’t reveal much about post-translational modifications. By merging several omics techniques together, the researchers were able to gain additional insights that connect the genome to function. For example, they linked certain KRAS mutations to the upregulation of specific glycoproteins. These glycoproteins mediate cell migration, invasion, and adhesion. They also appear to protect neoplastic cells from a type of programmed cell death that occurs when cells detach from the extracellular matrix and that plays a role preventing cancer metastasis.
The connection between KRAS and these glycoproteins provides a potential point of therapeutic intervention. While KRAS itself is difficult to drug, it may be possible to develop monoclonal antibodies against these glycoproteins. In patients with KRAS mutations that upregulate these glycoproteins, such monoclonal antibodies could work in conjunction with first-line chemotherapies.
Addressing difficulties in treating PDAC: Immune evasion and biomarkers
One reason it is difficult to treat PDAC with immunotherapies is that immune cells typically have difficulty penetrating pancreatic tumors. Using a combination of transcriptomic and phosphoproteomic approaches, Cao et al. 2021 discovered that “immune-cold” tumors had reduced expression of endothelial adhesion proteins and upregulation of a variety of pathways that might make the tumors impermeable to small molecules and immune cells. Such pathways included:
- VEGF – Involved in endothelial cell remodeling during tumorigenesis
- Hypoxia – Involved in endothelial cell remodeling during tumorigenesis
- Glycolysis – Involved in the generation of ATP (most cancer cells rely on glycolysis for proliferation)
- Cell junctions – Involved in regulating the permeability of endothelial cells to small molecules and immune cells
Targeting the above processes pharmacologically may make it easier for immune cells to invade pancreatic tumors and prevent tumor growth.
Another reason that PDAC is difficult to treat is that it is often diagnosed at an advanced stage. Therefore, new biomarkers could enable earlier diagnoses. To work towards this goal, Cao et al. 2021 identified 12 secreted proteins that are over 2-fold upregulated in PDAC. They also found thousands of protein phosphorylation sites and N-linked glycosites that were increased in abundance in PDAC. These may one day make great biomarkers for the early identification of the disease and may therefore result in many saved lives.
The importance of a multiomics approach in understanding cancer
This study highlights the importance of a multiomics approach in understanding disease. Many disease biomarkers and affected pathways cannot be identified based on genomics or transcriptomics alone and this study strongly reinforced that point. With their multiomics approach, the researchers identified new links between genomic alterations and their impact on the proteome, revealed various pathways involved in PDAC immune evasion, and discovered potential biomarkers for the early detection of PDAC.
Next-generation proteomics technologies, such as the NautilusTM Proteome Analysis Platform, are designed to reveal even more biological links in cancer. They aim to give researchers the ability to rapidly improve their understanding of cancer and other diseases by identifying up to 95% of proteins in a sample regardless of abundance. The wide accessibility of such technologies will hopefully reveal new biomarkers and drug targets that will vastly improve our ability to diagnose and treat pancreatic cancer and many other diseases.
Discover additional ways next-generation proteomics can fuel biology research in our applications of proteomics blog posts.
MORE ARTICLES