In this series, we discuss 3 types of research that can be accelerated by advanced proteomics technologies. This is far from an inclusive list and focuses on non-clinical research with a nod to clinical applications that can result from these research areas. As we get accessible, high-throughput proteomics technologies into the hands of researchers around the world, they’ll develop myriad applications that will reveal much more about the inner-workings of living things than we ever thought possible.
Enhancing our understanding of biological function and dysfunction through the identification of proteoforms
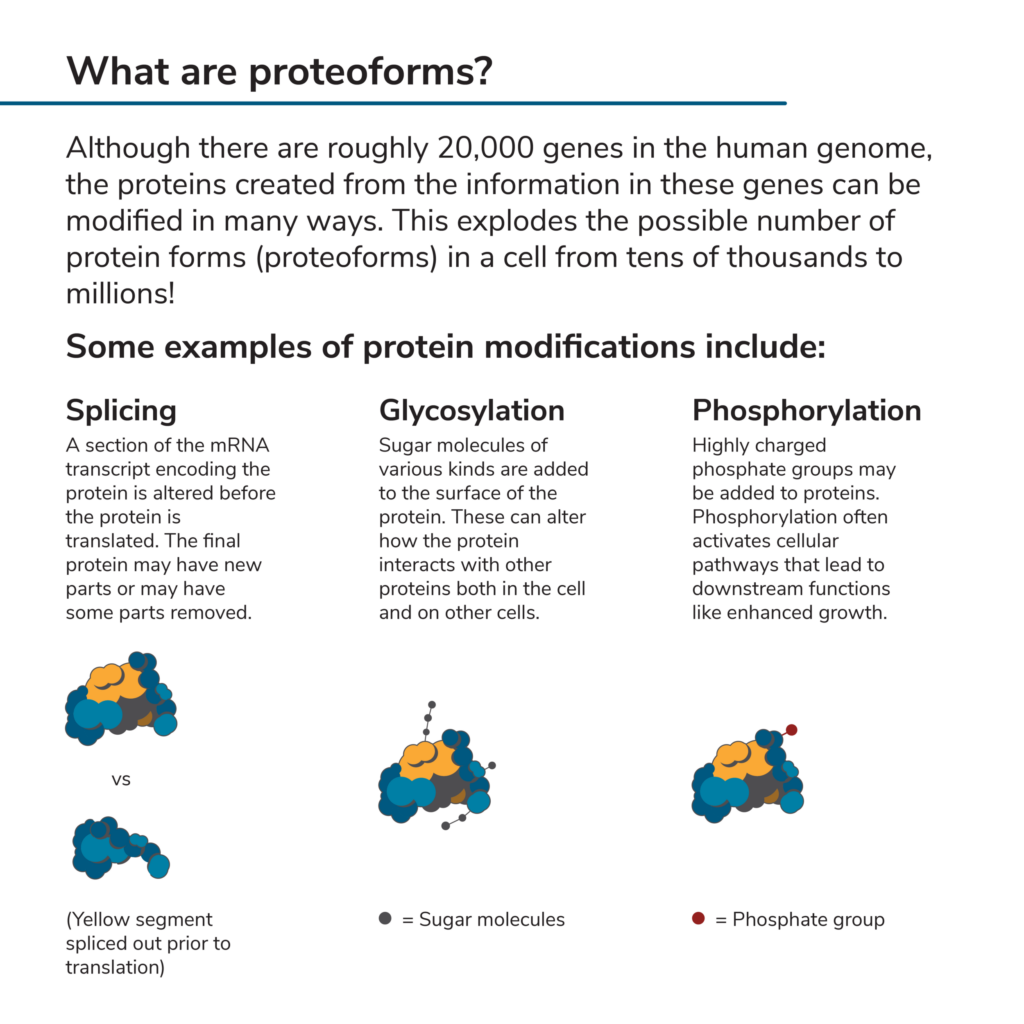
You may have heard that there are roughly 20,000 genes in the human genome. One might assume that, since genes encode proteins, there can be no more than 20,000 different proteins in any given cell. However, this is far from the case. In the process of decoding the information in a gene and using it to produce a protein, that information can be contorted in a variety of ways that leave out some parts of the original gene, start the decoding processes at different places in the gene, or even edit small portions of the information. In addition, once this information is used to create a protein, the protein itself can be modified with a variety of attachments such as chemicals and sugars. All this rearranging and modification explodes the number of possible “proteoforms” that could theoretically be produced in a cell from 20,000 to millions (Aebersold et al 2018).
The modifications found on proteoforms can change the underlying protein’s function or even lead to its destruction. Indeed, the precise mix of proteoforms in a cell can have great impacts on cell processes, organ function, and total body health. Thus, knowing more about the proteoforms present in any given healthy or diseased cell may give scientists great insights into how these cells operate.
Unfortunately, most proteomics technologies used today can only see a small fraction of proteoforms. Reasons why include:
Low sensitivity limits the proteoforms that can be observed
There are many different ways proteins can be modified to create a wide array of proteoforms. However, for any given cell, it’s possible that only a few proteoforms will be present in high abundance. There may be low abundance proteoforms that have functional impacts on the cell, but they will be hard to see as the more abundant proteoforms drown out their signals on traditional proteomics platforms. In addition, as most proteomics platforms look at groups of cells as opposed to individual cells, if one cell is dominated by a proteoform that is rare in other cells, that proteoform will be difficult to see.
Limitations due to protein fragmentation
On traditional proteomics platforms (e.g. mass spectrometers), full-length proteins are fragmented into small chunks. These small chunks are identified, and analysis software pieces together full proteins from the identified chunks. However, not all the chunks that make up the full protein will actually be identified. Some chunks will be left out of the analysis and those chunks could have modifications that are invisible to standard proteomics technologies.
Limited means of detection
Traditional proteomics platforms generally detect the mass of protein fragments in order to identify them. They look to see how many fragments of various weights are present and, by plotting abundance vs mass, they generate so-called “spectra” which are essentially signatures of different fragments. However, some fragments can generate very similar spectra and, without reference samples to distinguish between them, it can be difficult to identify fragments with never-before-seen modifications.
The benefit of detecting new proteoforms
There are many unknowns when it comes to the world of proteoforms. Scientists have made theoretical predictions about the number of proteoforms that could possibly exist, but it is not at all clear what fraction of these proteoforms are actually made, how they might be distributed across cells, and what functional consequences they have. Nonetheless, we do know that some protein modifications are highly consequential. For example, we know that an overabundance of certain types of modifications to proteins that scaffold DNA are associated with poor outcomes in cancer (Nebbioso et al 2018). If scientists can identify more proteoforms in particular cells, they can more confidently associate them with various states of health and disease. Some may even become biomarkers that indicate when a person has a particular disease and others may be targeted by novel drugs.
Enhancing proteoform detection with the Nautilus Proteome Analysis Platform
At Nautilus, we’re developing a proteomics platform that makes it easier to identify a much wider variety of proteoforms. Some of the ways the platform enhances proteoform identification include:
- The platform is designed to analyze individual proteins in isolation. This is expected to give the platform higher sensitivity than standard proteomics platforms and thus more potential to detect relatively rare proteoforms.
- The platform is designed to analyze full-length proteins. Thus, it is theoretically possible to identify modifications across the entire length of a protein.
- The platform is designed to use “probes” that detect particular sequences of protein building blocks in full-length proteins. These probes can be designed to detect building blocks that have been modified in a variety of ways and thereby detect various proteoforms.
By getting a more in-depth view of the proteoforms that exist across cells, we hope to take the first step toward understanding how these proteoforms impact health and disease. Once scientists observe differences in proteoform abundance across cells, they can investigate whether those differences are functionally significant, but the first step is seeing what proteoforms actually exist. We expect our platform to bring us a long way toward accomplishing this key step.
In the next post in this series, we discuss how proteomics to help researchers understand the molecular underpinnings of cell identity.
Want to read the full series now? Download the “Accelerating basic research with proteomics” white paper here.
MORE ARTICLES



