Nautilus™ Proteome Analysis Platform
Iterative Mapping is designed to achieve deep biological insights that were previously inaccessible
Iterative Mapping
Iterative Mapping enables simultaneous, single-molecule analysis of billions of proteins and proteoforms with high accuracy, precision, and industry-leading reproducibility. It is designed to support deeper biological understanding, novel biomarker discovery, and the advancement of precision medicine.
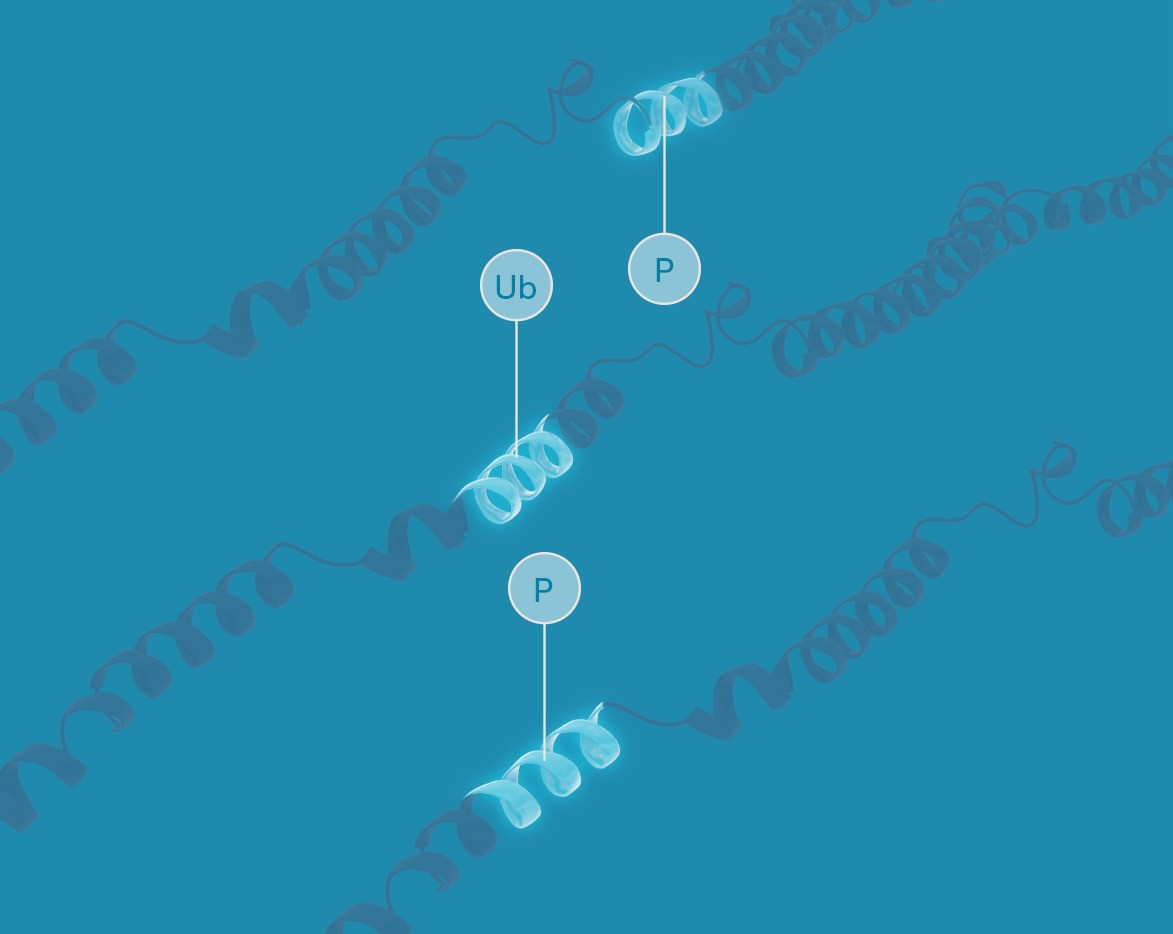
Targeted proteoform analysis
Targeted proteoform analyses provide in-depth views of individual protein molecules. We employ Iterative Mapping of proteoforms to identify targeted sets of modifications and isoform-specific sequences on full-length, single-molecule proteins. These analyses are designed to enable researchers to determine the biological impacts of the full patterns of modification found on proteoforms.
Advantages of Iterative Mapping
Comprehensive proteomic analysis
Iterative Mapping is conducted on massive arrays designed to capture the full proteome while quantifying proteins and proteoforms at the single-molecule level, achieving unprecedented coverage and detail.
Learn more
Comprehensive proteomic analysis
Multi-affinity probes enables quantification of 1,000s of unique proteins providing high quality data throughput per sample.
Accuracy
Many single-molecule probing events are designed to provide high confidence in protein and proteoform identifications. IDs are simply counted to deliver accurate abundance measurements.
Learn more
Accuracy
Up to 10 billion landing pads per sample provide up to 9 orders of magnitude dynamic range and limit the need for depleting high abundance proteins.
Reproducibility
Single-molecule accuracy and precision are designed to combine to provide high reproducibility that enables you to trust your data and focus on insights instead of technical noise.
Learn more
Reproducibility
Single-molecule counting provides the highest sensitivity for detection and quantification of proteins and proteoforms.
Standardization and scalability
Single-molecule Iterative Mapping underlies all applications of the platform. Ultra-dense single-molecule arrays are designed to scale to the full proteome while streamlined workflows enable implementation in any lab.
Learn more
Standardization and scalability
The combined use of hundreds of multi-affinity probes, a machine learning algorithm that parses binding data into high probability protein identifications, and automated workflows minimizes errors and improves reproducibility.
AI-readiness
High quality, simple, scalable, and direct measurements of biology make Nautilus data well-suited for AI integration and multiomic analysis.
Learn more
AI-readiness
Delivering digital counts with a simple workflow avoids lengthy, time-consuming data analysis that requires specialized expertise.
Integrated workflow from sample prep to protein quantification

Protein library preparation and deposition
Functionalize your proteins to capture and immobilize them on hyper-dense single-molecule arrays.
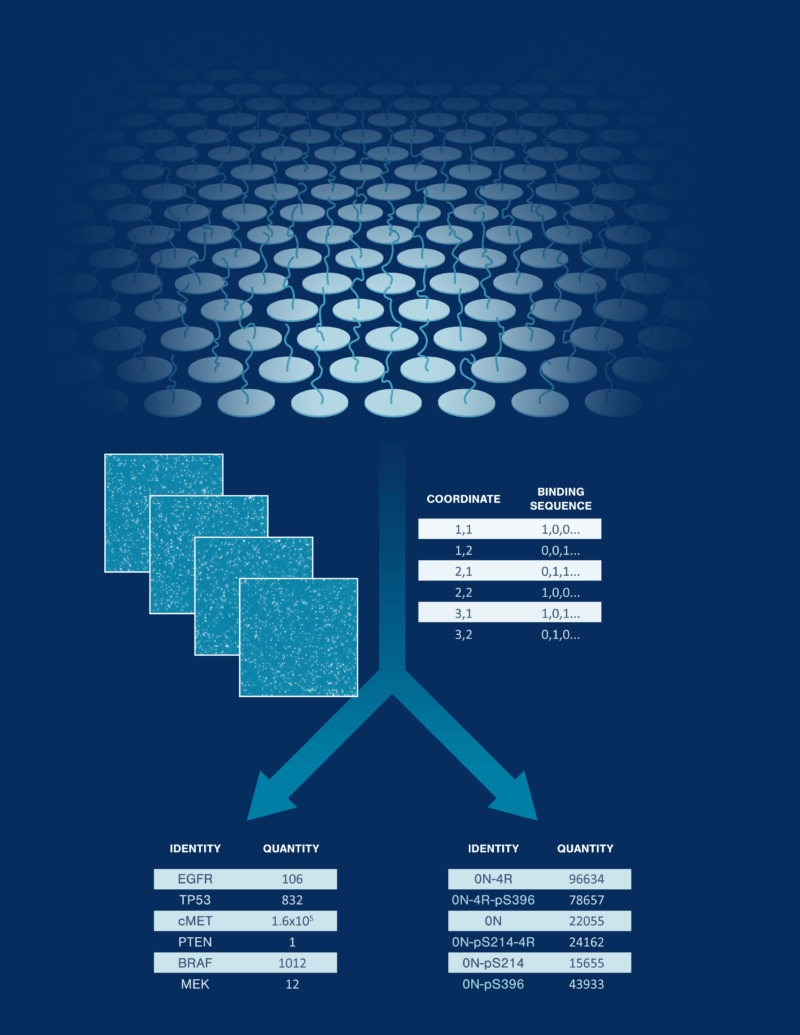
Repeated probing
Iteratively interrogate every immobilized protein using probes designed to bind single-molecule features indicative of protein or proteoform identity. Machine learning-powered algorithms convert the probe binding patterns into confident protein and proteoform identifications that are summed for robust quantification.
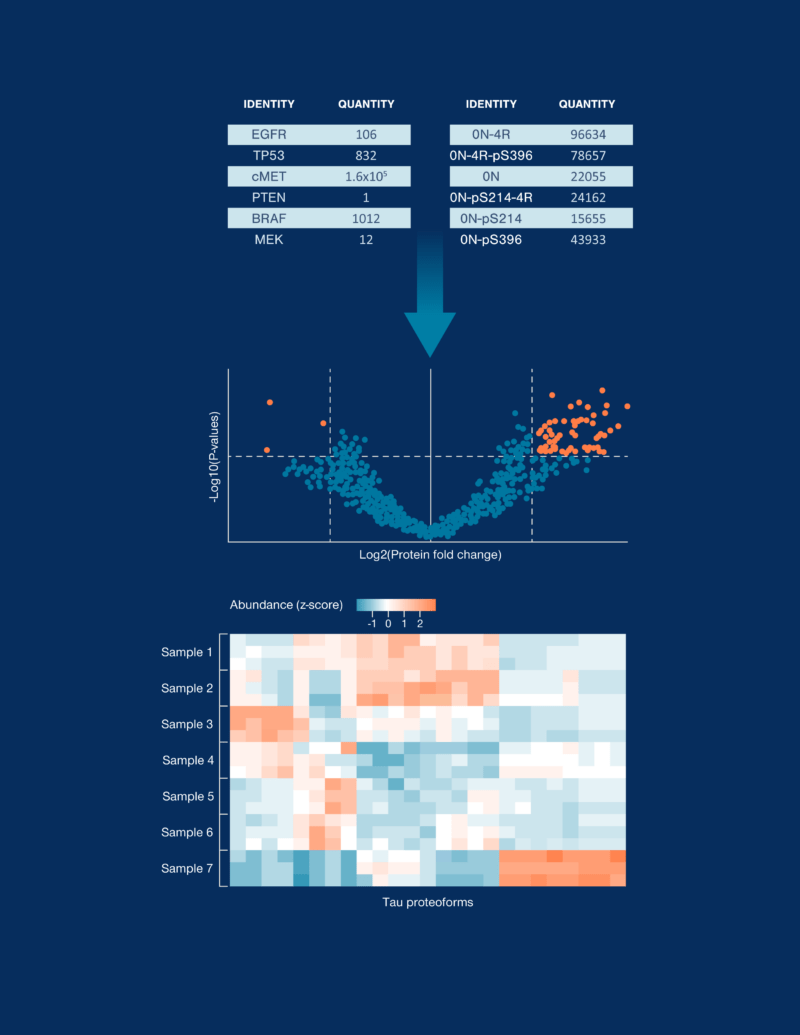
Comprehensive proteomic analysis
The platform generates a simple output format with protein or proteoform identities and counts. Data can be further analyzed in Nautilus’ cloud portal to derive biological insights.
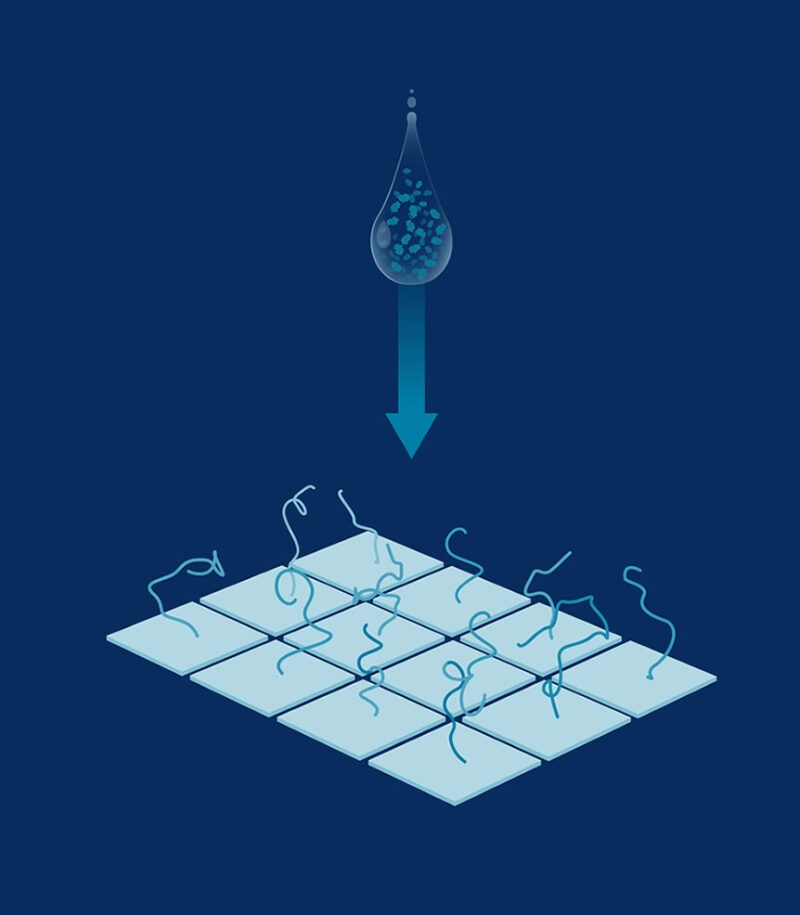
Protein library preparation
Proteins are functionalized to produce a library of single-molecules on a hyper-dense nanoarray.
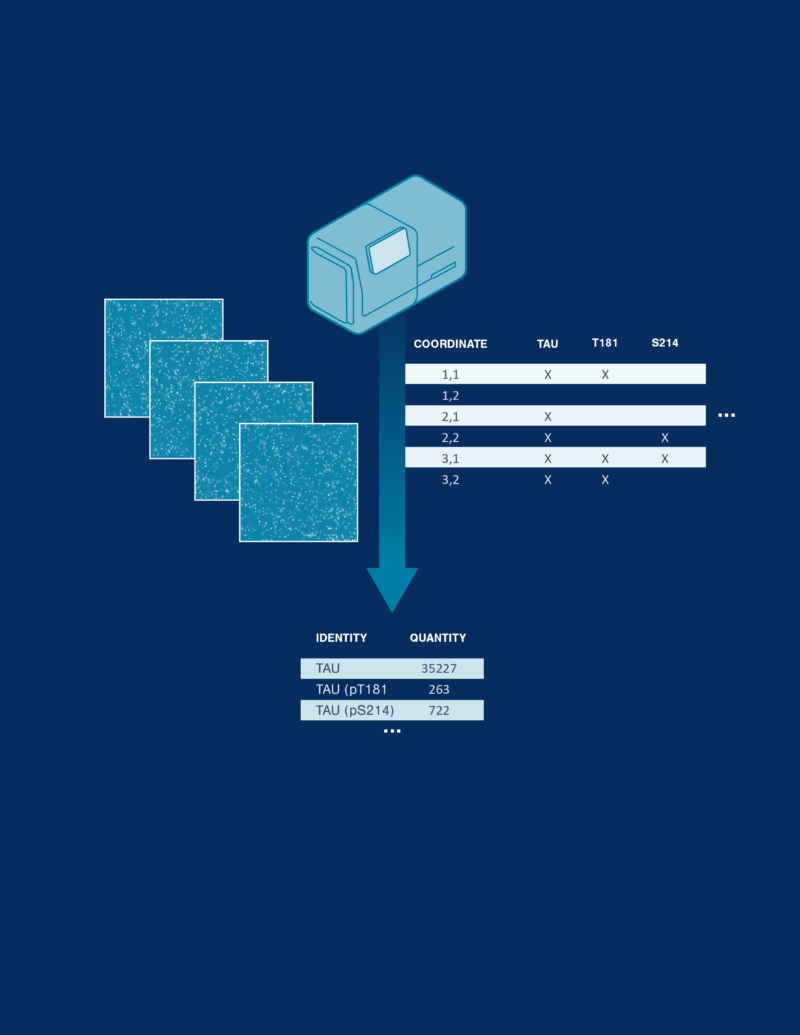
Proteoform identification and quantification
Targeted probes specific to proteoforms of interest are used to identify proteoforms on the platform. Quantification is performed by counting how many molecules of each proteoform are observed.
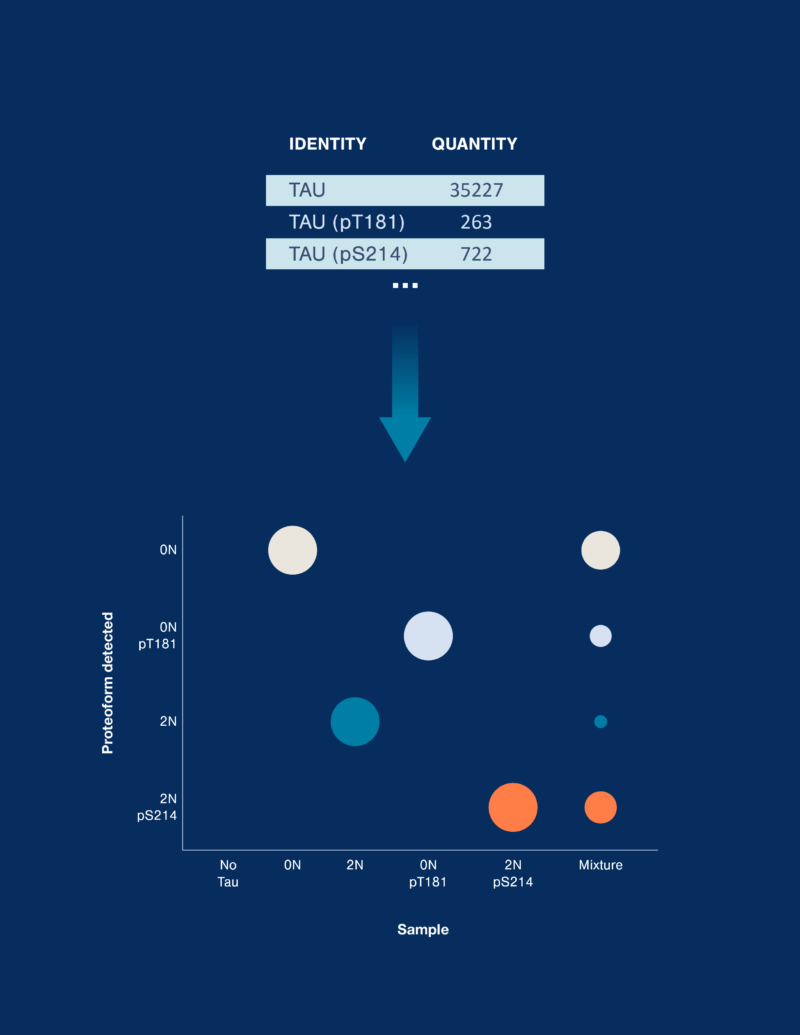
Data interpretation
Quantified proteoforms are analyzed with a variety of visualization tools to reveal new insights.
One method enabling endless applications for full proteome coverage with exceptional detail
Explore uncharted biology

Revolutionize proteomics and be one of the first to apply Iterative Mapping in your research

