Iterative Mapping of proteoforms may reveal novel biology and new drug targets
July 1, 2025
If researchers gain more knowledge of the proteoforms that underlie biology, they may be able to create more effective therapeutics and actionable diagnostics.
This is because proteoforms may reveal the in-depth molecular mechanisms underlying disease pathologies. Briefly, proteoforms are variants of proteins defined by their combinations of genetic variation, alternative splicing, post-translational modification, and any other alterations. Individual proteoforms derived from the same gene can have altered levels of activity, altered localization, and may even participate in entirely different biological processes. Ultimately, proteoforms are the functional variants of proteins found in biology. As such, proteoforms may have unique roles in certain biological processes and pathological phenotypes. They may also make ideal targets for next-generation therapeutics.
In our recent preprint, we use Iterative Mapping on the NautilusTM Proteome Analysis Platform to explore tau proteoforms and their roles in Alzheimer’s Disease and Related Dementias (ADRD) – this exploration serves as an exemplar for why proteoform analysis has the potential to accelerate biological discovery and therapeutic development. Over the next month, we’ll dive further into the data from the preprint and discuss:
- Metrics demonstrating the benefits of Iterative Mapping on the Nautilus Proteome Analysis Platform (This post).
- The new kinds of analysis Iterative Mapping of proteoforms enables (Post 2).
- How Iterative Mapping answers key questions about tau proteoform biology (Post 3).
- How our preliminary tau proteoform data demonstrates the potential impact of proteoform-level analysis on the development of next-generation therapeutics and biomarkers for not just ADRD, but all diseases (Post 4).
Read our preprint covering Iterative Mapping of proteoforms
Explore the quantitative benefits of Iterative Mapping of proteoforms in our Tech Note
Iterative Mapping as a means of measuring proteoforms
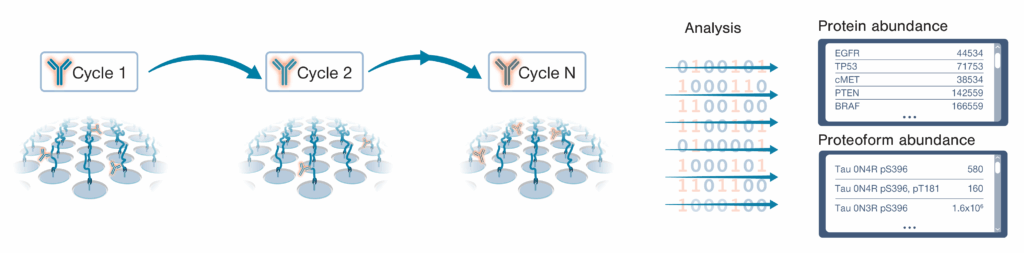
The NautilusTM Proteome Analysis Platform employs a method called “Iterative Mapping” to measure proteins in broadscale proteomic analyses and proteoforms in targeted proteoform analyses. In Iterative Mapping, millions to billions of intact, denatured protein molecules are bound to nano-fabricated flow cells at distinct coordinates. Then, the proteins are interrogated with fluorescently labeled probes designed to bind either short amino acid sequences (~3 amino acids) in broadscale proteomic analysis or proteoform features such as post translational modifications and isoform-specific sequences in targeted proteoform analysis. These probes are iteratively introduced to the flow cells (one probe per cycle). Then, machine learning-powered algorithms use observed binding patterns to identify each protein or proteoform at the single-molecule level, and identifications are summed to measure abundance (Figure 1).
Figure 1: Iterative Mapping for quantitative, single-molecule proteome and proteoform analysis.
In our preprint, we share key metrics demonstrating the benefits of Iterative Mapping of proteoforms. We assessed the following benefits of the method:
- Accuracy – It measures expected proteoforms at their expected abundances.
- Sensitivity – The observed limit of quantification, ~0.1% relative abundance, is expected to be sufficient to discern small abundance differences between members of a given proteoform family.
- Dynamic range – It can measure high and low-abundance proteoforms over ~3 orders of magnitude relative abundance.
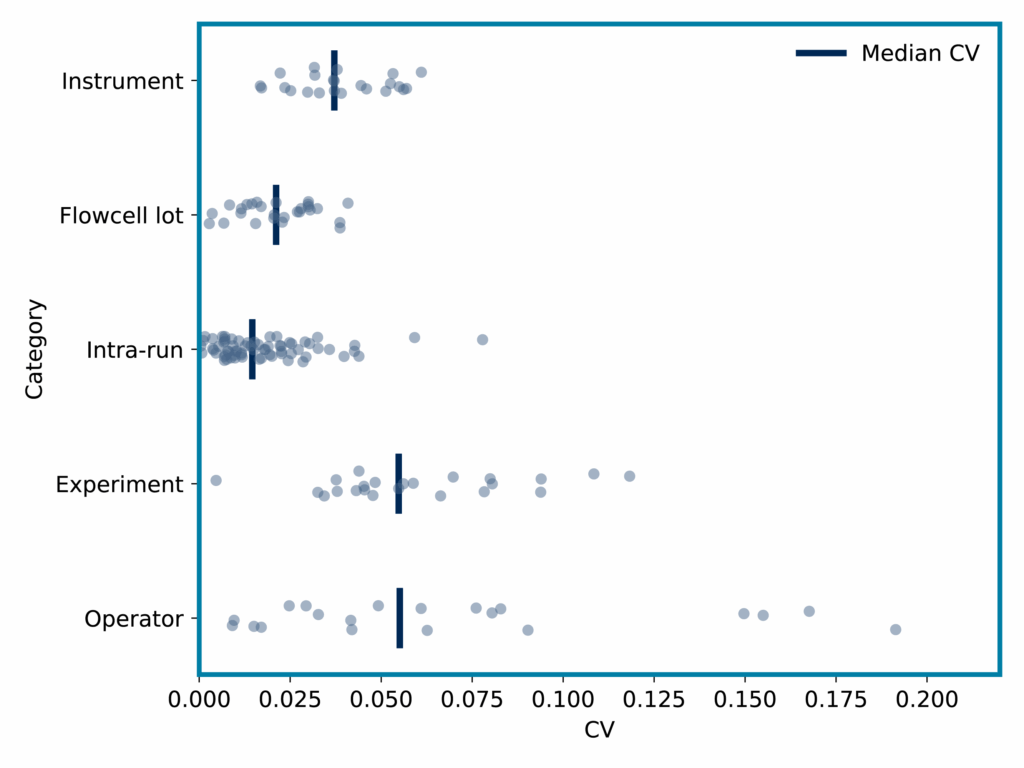
- Reproducibility – Median CVs were less than 6% across many sources of variability (Figure 2).
Figure 2: Iterative Mapping of proteoforms CVs across various sources of variability.
Tau proteoforms are exemplars for the importance of proteoform research
Three AD treatments that modestly slow cognitive decline were recently approved and all target the amyloid beta protein. It is a primary component of the so-called “amyloid plaques” that form outside neurons in the brains of AD patients.
Potentially just as important as amyloid plaques are the neurofibrillary tangles (NFTs) formed by the aggregation of tau inside neurons. While amyloid plaques can appear in the brain many years before cognitive symptoms of AD, tau aggregation is correlated with cognitive decline. What causes NFTs to form is not clear, but researchers have shown that tau becomes progressively more modified as AD progresses, and that particular modifications such as pT217 can be indicators of disease.
Beyond AD, tau is known to play a role in a variety of other neurodegenerative diseases collectively called “tauopathies.” Thus, learning more about this protein and how its proteoforms impact disease may enable the development of therapeutics targeting them. This could be a boon for people suffering from a wide variety of neurodegenerative conditions. Many hope that targeting tau will do much more than modestly slow cognitive decline in AD.
Tau and neurodegeneration – What’s known?
Tau is a complex protein expressed as 6 different isoforms (Goedert et al., 1989) in the central nervous system (CNS) (Reviewed in Alquezar et al., 2021) and 1 additional isoform (big tau) in the peripheral nervous system (Fischer and Baas 2021). Although there has been some evidence that big tau is neuroprotective (see preprint by Chung et al., 2024), the 6 CNS isoforms are the most studied in neurodegeneration and we focus on them in the preprint.
As Translating Proteomics podcast guest and AD expert Sarah DeVos eloquently stated in a discussion about AD, “Anything that’s -ated, tau has it at some point.” In other words, tau is highly modified. It undergoes phosphorylation, acetylation, methylation, ubiquitination, and more at many sites. Furthermore, bottom-up mass spectrometry has demonstrated the relevance of these modifications showing that:
- Certain tau isoforms and modifications gain prominence as AD progresses (Wesseling et al., 2020).
- Certain tau modifications are more prominently associated with the seeding of tau aggregates (Dujardin et al., 2020).
- Certain tau isoforms and modifications may be more prominently associated with different tauopathies (Kyalu Ngoie Zola et al., 2023).
However, because bottom-up mass spectrometry requires digestion of proteins into peptides before analysis, it is not clear what combinations of isoforms and modifications make up the tau proteoforms found in any given disease, at any given stage of disease, or in any given tau fraction. In addition, while it is known that some modifications are more prominent at later stages of disease, it is not clear if there is a prescribed order and timing to the creation of different tau proteoforms. Certain combinations of isoforms and modifications may preclude, enable, or even drive the creation of other combinations, but bottom-up mass spectrometry is not, as proteoform expert Neil Kelleher says, a “proteoform-aware” technology and cannot reveal this information.
Proteoform-aware technologies may reveal opportunities for drug development
Proteoform-aware technologies are critical because they may reveal the most pathogenic proteoforms of a protein like tau, what proteoforms precede their creation, and what processes enable their creation. With this information in-hand, researchers could potentially design diagnostics to detect pathogenic proteoforms and drugs that prevent their creation or clear them from the body.
For instance, if a theoretical researcher discovered the following:
- That a quadruply-modified tau proteoform appeared just before the onset of Alzheimer’s symptoms.
- That treating model systems with this proteoform strongly induced the molecular and physiological hallmarks of cognitive decline.
- And that there was a defined path for the creation of this proteoform.
They might:
- Develop antibodies that remove this proteoform from the brain to potentially prevent its physiological impacts.
- Develop small molecules that specifically inhibit the activity of modification enzymes that create this proteoform.
- Develop small molecules that inhibit interactions between the proteoforms that precede it and the enzymes that modify them.
Without proteoform-aware technologies, these potential paths for drug development would be invisible to researchers. With proteoform-aware technologies, many new therapeutic opportunities may present themselves.
The power of Iterative Mapping of proteoforms
In our preprint, we used Iterative Mapping to measure tau proteoforms across a variety of model systems and ADRD patient samples. While these are preliminary studies, they demonstrate the accuracy, sensitivity, dynamic range, and reproducibility of this novel, single-molecule method. Furthermore, our results show:
- Different model systems have different mixtures of tau proteoforms.
- Tau proteoform measurements differentiate cognitively normal samples from ADRD samples.
- There may be an order to the modifications that produce tau proteoforms.
- Severe ADRD samples may contain unique tau proteoforms.
We will explore these results in depth in a series of blog posts published over the next month. Critically, Iterative Mapping can be applied to any proteoforms so long as the probes necessary to identify their isoforms, post translational modifications, and other alterations are available.
If you’d like more information now, you can:
MORE ARTICLES