Creating hyper-dense protein arrays for single-molecule proteomics

Tyler Ford
January 29, 2024
Single-molecule proteomics interrogates the proteome at the highest sensitivity possible by discretely analyzing individual protein molecules. Yet, the sheer scale of the proteome makes single-molecule proteomics difficult. Most single-molecule protein analysis technologies today can’t analyze the entire proteome of a sample at once, making proteome-scale efforts laborious, time-consuming, and impractical.
On the NautilusTM Proteomic Analysis Platform, we leverage hyper-dense protein arrays with billions of landing pads that each isolate single protein molecules. The sheer amount of landing pads is designed to enable researchers to quantify both high- and low-abundance proteins in the proteome of any sample. With this technological advancement we aim to make broadscale discovery proteomics accessible to many more researchers and thereby spark a proteomics revolution.
For a more technical description of the topics discussed below, check out our tech note or dive even deeper with our white paper on our hyper-dense arrays.
Designing arrays for single-molecule proteomics
Combining a few key techniques enables us to create hyper-dense single-molecule protein arrays for next-generation proteomic analysis.
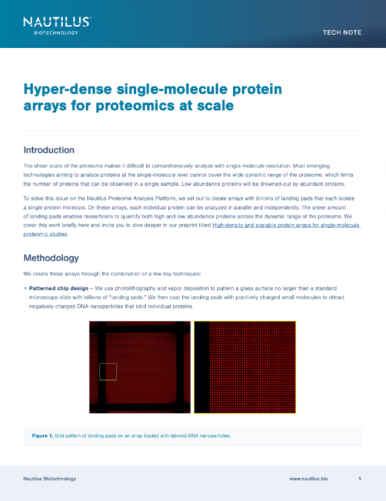
Patterned chip design — Protein arrays are patterned with billions of “landing pads,” that are designed to bind individual protein molecules.
Custom nanoparticle production — Our hyper-dense protein arrays leverage nanoparticles that bind to individual protein molecules. Each nanoparticle can capture one protein and only one nanoparticle-protein conjugate can fit on a landing pad. This enables us to distinguish individual protein molecules for single-molecule proteomics.
Click chemistry — Single protein molecules attach to nanoparticles in a simple, scalable chemical reaction.
Testing hyper-dense single-molecule arrays
Conducting single-molecule proteomics with the Nautilus Proteomic Analysis Platform depends on isolating single proteins, each attached to a nanoparticle on each individual landing pad. In our tests, very few landing pads (less than two percent) captured more than one protein. This single-molecule separation enables us to interrogate the full proteome at the single-molecule level for comprehensive single-molecule proteomics.
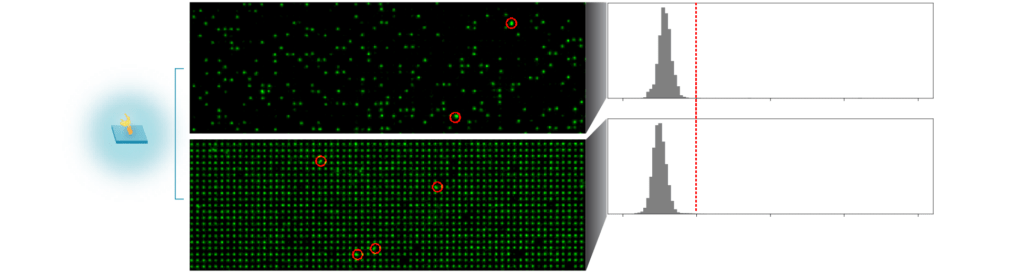
Our protein array with fluorescently labeled protein-nanoparticle conjugates attached to individual landing pads. Circles show rare instances of multiple proteins on one landing pad. The chart on the right shows higher-than-average signal intensity (indicating more than one protein) is very rare.
Unlocking single-molecule proteomics
Tests of the hyper-dense protein arrays that underpin the Nautilus Proteome Analysis Platform show that we can confidently isolate billions of proteins and conduct single-molecule protein analysis. This technology brings us closer to analyzing all the proteins in the proteome and makes single-molecule proteomics possible.
Learn more about the Nautilus Platform here.
MORE ARTICLES